statsmodels.tsa.arima_model.ARMAResults¶
-
class
statsmodels.tsa.arima_model.ARMAResults(model, params, normalized_cov_params=None, scale=1.0)[source]¶ Class to hold results from fitting an ARMA model.
Parameters: model : ARMA instance
The fitted model instance
params : array
Fitted parameters
normalized_cov_params : array, optional
The normalized variance covariance matrix
scale : float, optional
Optional argument to scale the variance covariance matrix.
Returns: Attributes
aic : float
Akaike Information Criterion
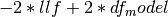 where df_model includes all AR parameters, MA parameters, constant
terms parameters on constant terms and the variance.
where df_model includes all AR parameters, MA parameters, constant
terms parameters on constant terms and the variance.arparams : array
The parameters associated with the AR coefficients in the model.
arroots : array
The roots of the AR coefficients are the solution to (1 - arparams[0]*z - arparams[1]*z**2 -...- arparams[p-1]*z**k_ar) = 0 Stability requires that the roots in modulus lie outside the unit circle.
bic : float
Bayes Information Criterion -2*llf + log(nobs)*df_model Where if the model is fit using conditional sum of squares, the number of observations nobs does not include the p pre-sample observations.
bse : array
The standard errors of the parameters. These are computed using the numerical Hessian.
df_model : array
The model degrees of freedom = k_exog + k_trend + k_ar + k_ma
df_resid : array
The residual degrees of freedom = nobs - df_model
fittedvalues : array
The predicted values of the model.
hqic : float
Hannan-Quinn Information Criterion -2*llf + 2*(df_model)*log(log(nobs)) Like bic if the model is fit using conditional sum of squares then the k_ar pre-sample observations are not counted in nobs.
k_ar : int
The number of AR coefficients in the model.
k_exog : int
The number of exogenous variables included in the model. Does not include the constant.
k_ma : int
The number of MA coefficients.
k_trend : int
This is 0 for no constant or 1 if a constant is included.
llf : float
The value of the log-likelihood function evaluated at params.
maparams : array
The value of the moving average coefficients.
maroots : array
The roots of the MA coefficients are the solution to (1 + maparams[0]*z + maparams[1]*z**2 + ... + maparams[q-1]*z**q) = 0 Stability requires that the roots in modules lie outside the unit circle.
model : ARMA instance
A reference to the model that was fit.
nobs : float
The number of observations used to fit the model. If the model is fit using exact maximum likelihood this is equal to the total number of observations, n_totobs. If the model is fit using conditional maximum likelihood this is equal to n_totobs - k_ar.
n_totobs : float
The total number of observations for endog. This includes all observations, even pre-sample values if the model is fit using css.
params : array
The parameters of the model. The order of variables is the trend coefficients and the k_exog exognous coefficients, then the k_ar AR coefficients, and finally the k_ma MA coefficients.
pvalues : array
The p-values associated with the t-values of the coefficients. Note that the coefficients are assumed to have a Student’s T distribution.
resid : array
The model residuals. If the model is fit using ‘mle’ then the residuals are created via the Kalman Filter. If the model is fit using ‘css’ then the residuals are obtained via scipy.signal.lfilter adjusted such that the first k_ma residuals are zero. These zero residuals are not returned.
scale : float
This is currently set to 1.0 and not used by the model or its results.
sigma2 : float
The variance of the residuals. If the model is fit by ‘css’, sigma2 = ssr/nobs, where ssr is the sum of squared residuals. If the model is fit by ‘mle’, then sigma2 = 1/nobs * sum(v**2 / F) where v is the one-step forecast error and F is the forecast error variance. See nobs for the difference in definitions depending on the fit.
Methods
aic()arfreq()Returns the frequency of the AR roots. arparams()arroots()bic()bse()conf_int([alpha, cols, method])Returns the confidence interval of the fitted parameters. cov_params()f_test(r_matrix[, cov_p, scale, invcov])Compute the F-test for a joint linear hypothesis. fittedvalues()forecast([steps, exog, alpha])Out-of-sample forecasts hqic()initialize(model, params, **kwd)llf()load(fname)load a pickle, (class method) mafreq()Returns the frequency of the MA roots. maparams()maroots()normalized_cov_params()plot_predict([start, end, exog, dynamic, ...])Plot forecasts predict([start, end, exog, dynamic])ARMA model in-sample and out-of-sample prediction pvalues()remove_data()remove data arrays, all nobs arrays from result and model resid()save(fname[, remove_data])save a pickle of this instance summary([alpha])Summarize the Model summary2([title, alpha, float_format])Experimental summary function for ARIMA Results t_test(r_matrix[, cov_p, scale, use_t])Compute a t-test for a each linear hypothesis of the form Rb = q tvalues()Return the t-statistic for a given parameter estimate. wald_test(r_matrix[, cov_p, scale, invcov, ...])Compute a Wald-test for a joint linear hypothesis. wald_test_terms([skip_single, ...])Compute a sequence of Wald tests for terms over multiple columns Methods
aic()arfreq()Returns the frequency of the AR roots. arparams()arroots()bic()bse()conf_int([alpha, cols, method])Returns the confidence interval of the fitted parameters. cov_params()f_test(r_matrix[, cov_p, scale, invcov])Compute the F-test for a joint linear hypothesis. fittedvalues()forecast([steps, exog, alpha])Out-of-sample forecasts hqic()initialize(model, params, **kwd)llf()load(fname)load a pickle, (class method) mafreq()Returns the frequency of the MA roots. maparams()maroots()normalized_cov_params()plot_predict([start, end, exog, dynamic, ...])Plot forecasts predict([start, end, exog, dynamic])ARMA model in-sample and out-of-sample prediction pvalues()remove_data()remove data arrays, all nobs arrays from result and model resid()save(fname[, remove_data])save a pickle of this instance summary([alpha])Summarize the Model summary2([title, alpha, float_format])Experimental summary function for ARIMA Results t_test(r_matrix[, cov_p, scale, use_t])Compute a t-test for a each linear hypothesis of the form Rb = q tvalues()Return the t-statistic for a given parameter estimate. wald_test(r_matrix[, cov_p, scale, invcov, ...])Compute a Wald-test for a joint linear hypothesis. wald_test_terms([skip_single, ...])Compute a sequence of Wald tests for terms over multiple columns Attributes
use_t
